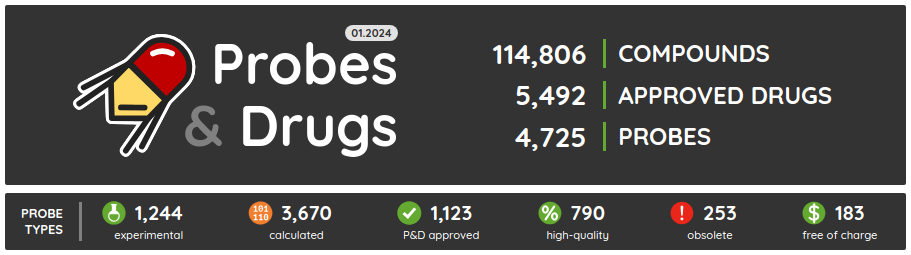
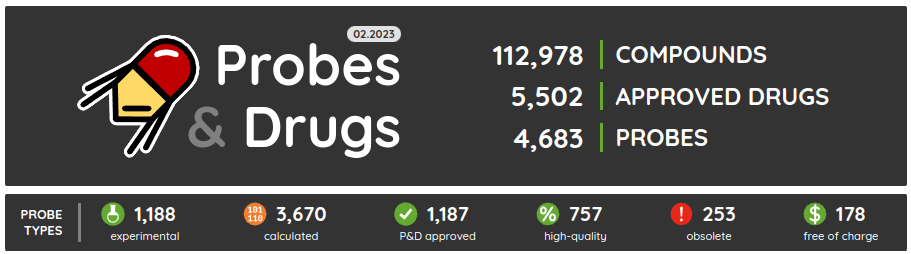
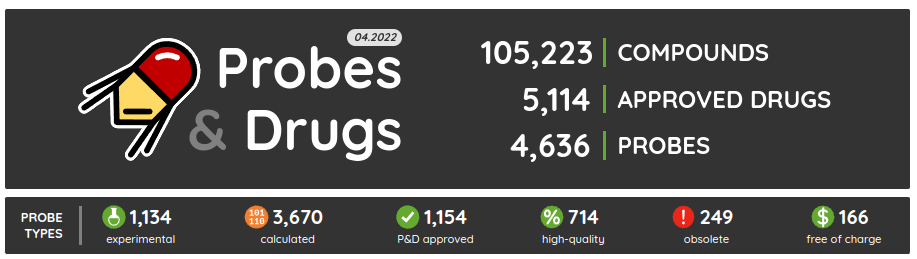
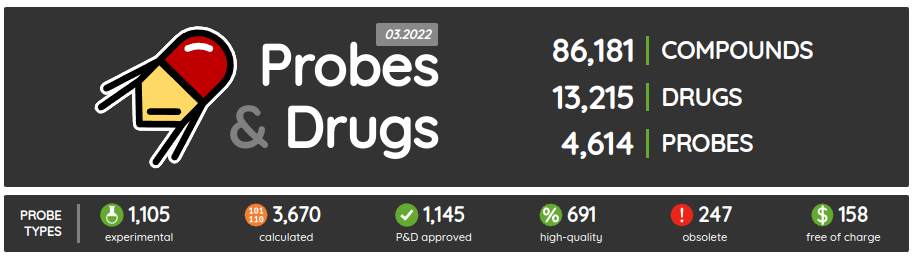
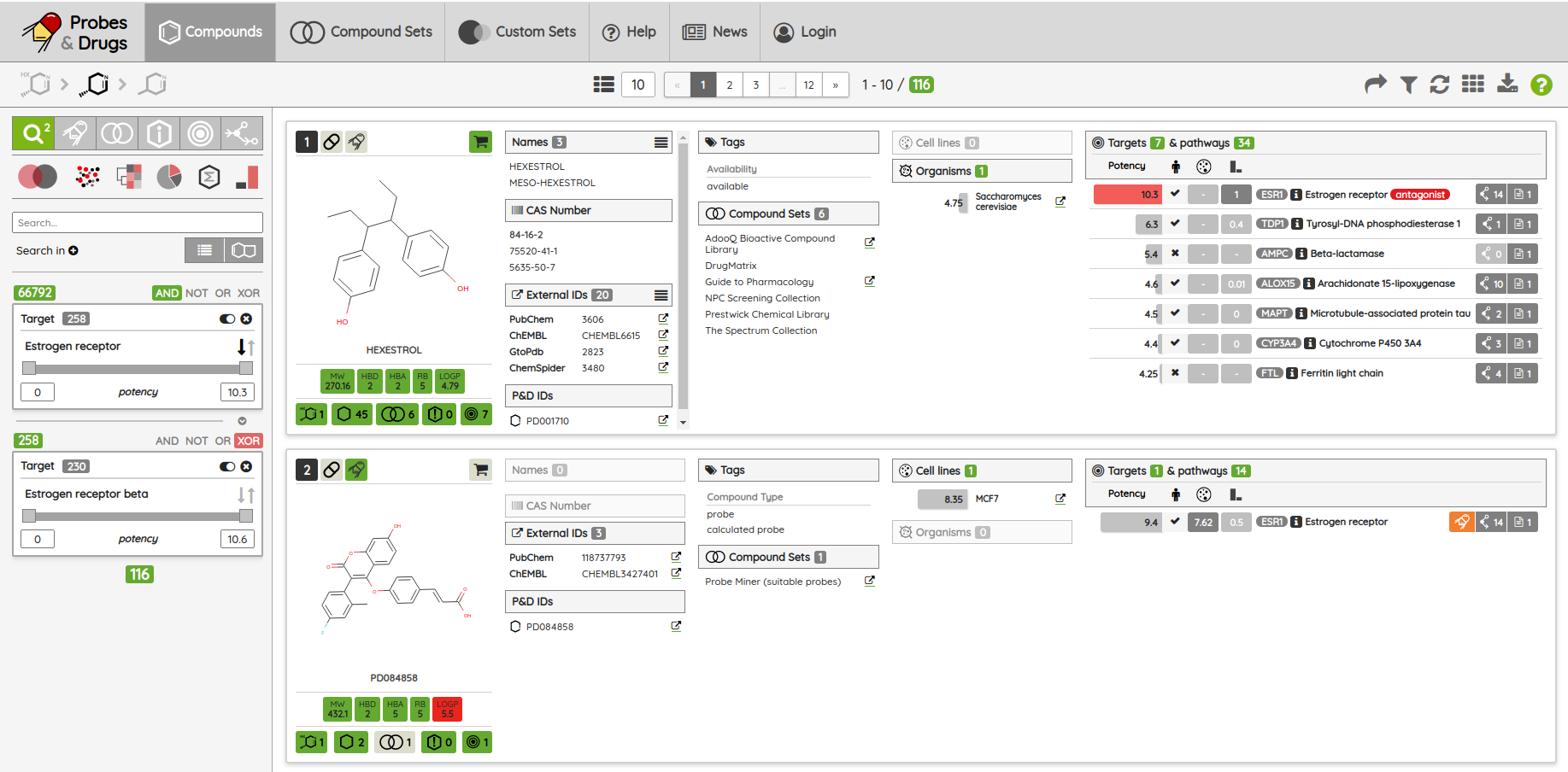
The first release of the Probes & Drugs this year - 01.2024 - is here with the new Concise Guide to Pharmacology 2023/24 set, withdrawn drugs from Withdrawn DB and a glossary to help users with the orientation in the nomenclature used on the portal. Also, all major data sources (ChEMBL, Guide to Pharmacology, BindingDB, DrugBank, Reactome and others) were updated to their latest versions as well as several of the live compound sets (such as compounds from Chemical probes.org, SGC/SGC Donated/opnMe chemical probes and others). Apart from the updated ones, we added three new compound sets: Concise Guide to Pharmacology 2023/24, Withdrawn 2.0 DB and BOC Sciences Bioactive Compound library.
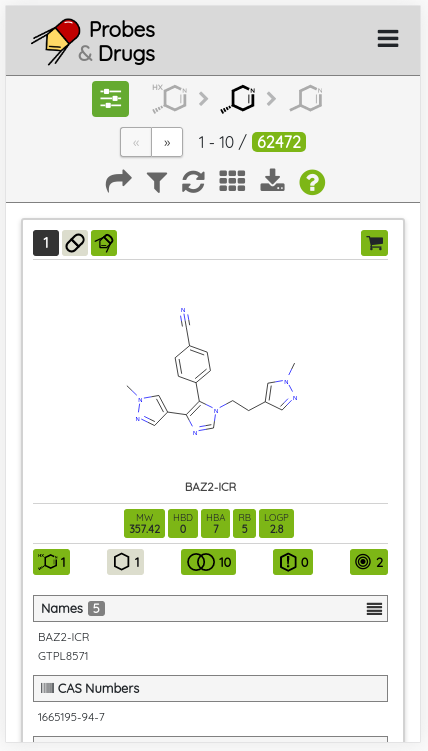
Concise Guide to Pharmacology 2023/24 (CGTOP) is a set of high-quality manually curated set of pharmacologically active compounds published and extracted from the biennial series of publications in the British Journal of Pharmacology. This compound set can be perceived as a selected subset of compounds from the Guide to Pharmacology and it is also one of the sets used for the selection of our High-quality chemical probes set. The latest CGTOP version comprises 2879 standardized compounds and contains 234 compounds not found in the previous version of the set.
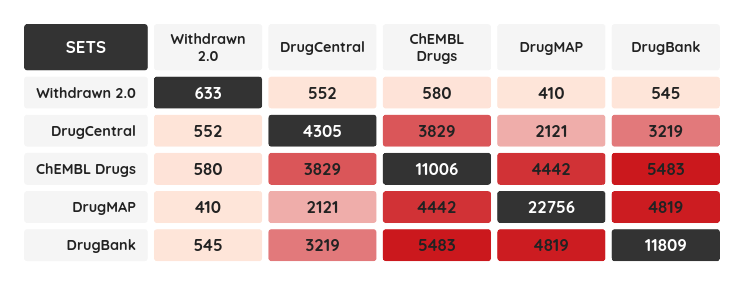
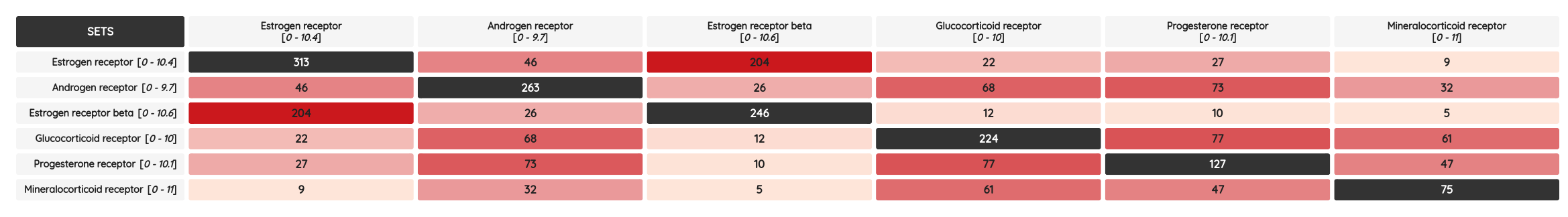
The Withdrawn 2.0 is a manually curated database of withdrawn drugs aiming to facilitate understanding the underlying mechanisms that lead to the withdrawal, not only listing withdrawal information but also important features such as toxicity information, side-effects, targets, ATC classes and more. The set comprises 633 withdrawn drugs and has a high overlap with other drug compound sets on the portal: ChEMBL, DrugBank, DrugCentral and DrugMap. The intersection of all these resources is 380 compounds and the highest one is with drugs from ChEMBL, 580 drugs. The lowest one, 410 drugs, is surprisingly with DrugMap - by far the largest drug set on the portal with almost 23k compounds. The intersection matrix for all mentioned resources can be found below.
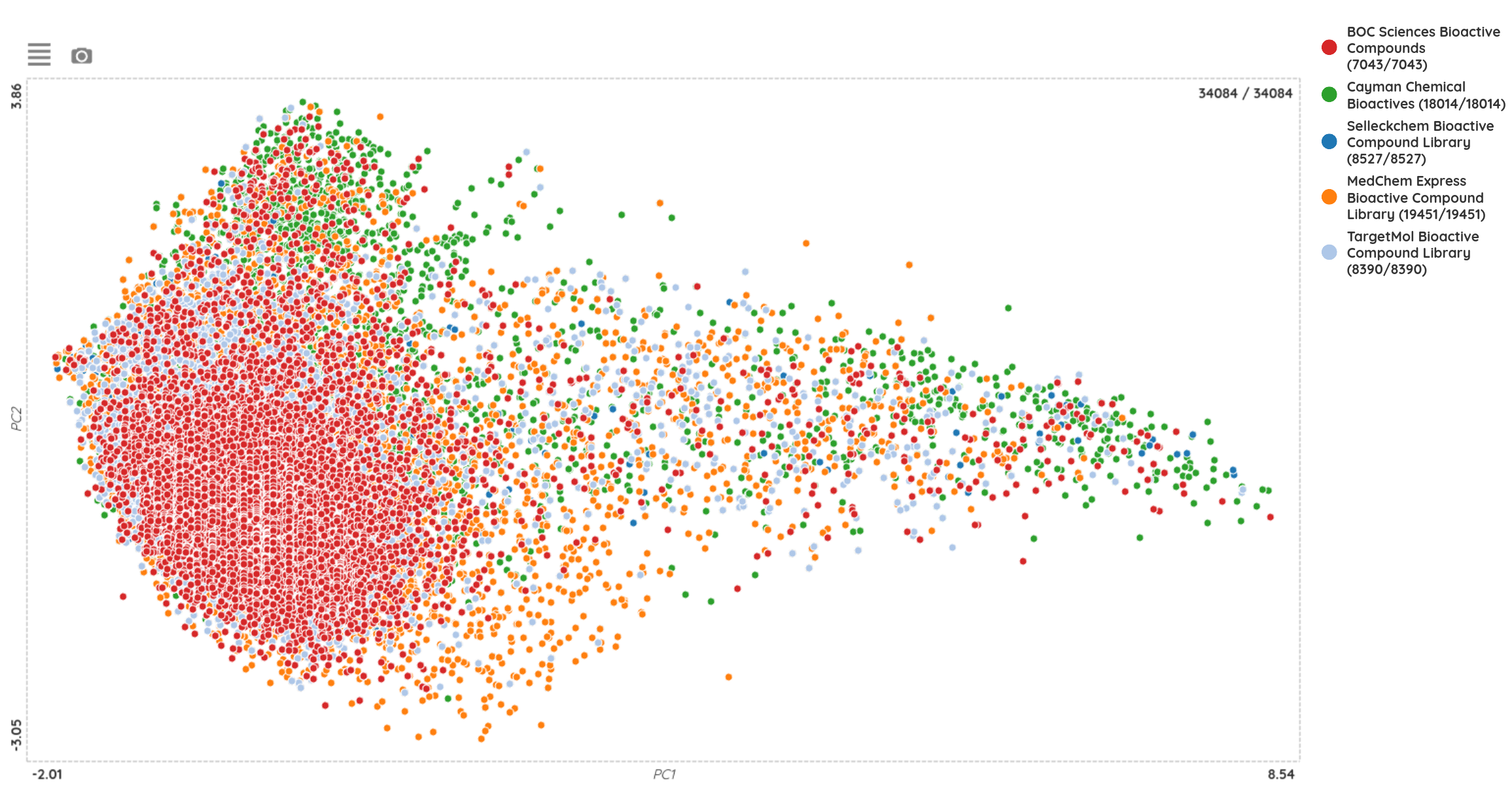
The last new set, the commercial bioactive compound library from BOC Sciences, is represented by 7,043 bioactive compounds. When compared with other up-to-date commercial bioactive compound sets (from Cayman, MedChem Express, SelleckChem and TargetMol), we can see that all of these sets contain unique compounds from some part, but all of them share more-or-less similar chemical space.
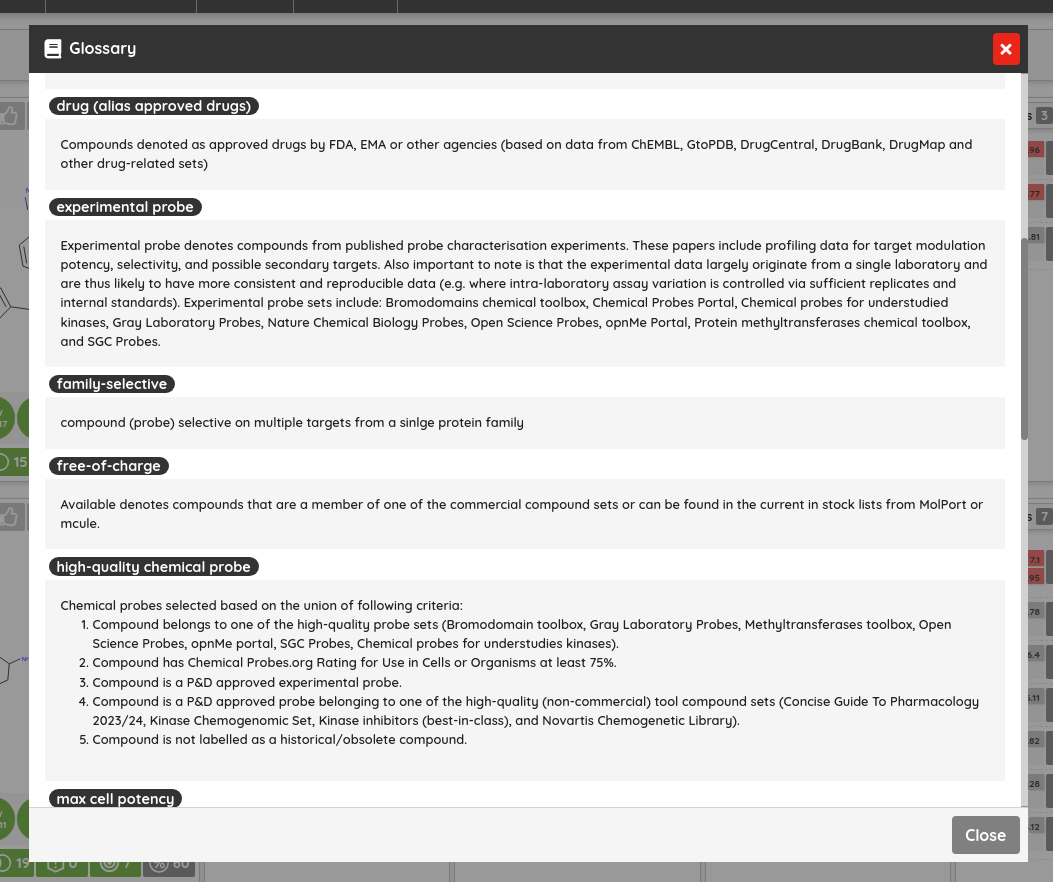
One of the new features that can be found on the portal from the current version is the glossary. The glossary contains various terms (tags, labels, keywords) used at the portal with their short explanation. It can be accessed by clicking on the book icon in the top right corner of the Compounds view. We hope that it will help new users to dive into P&D and better understand the data/compounds they are working with.
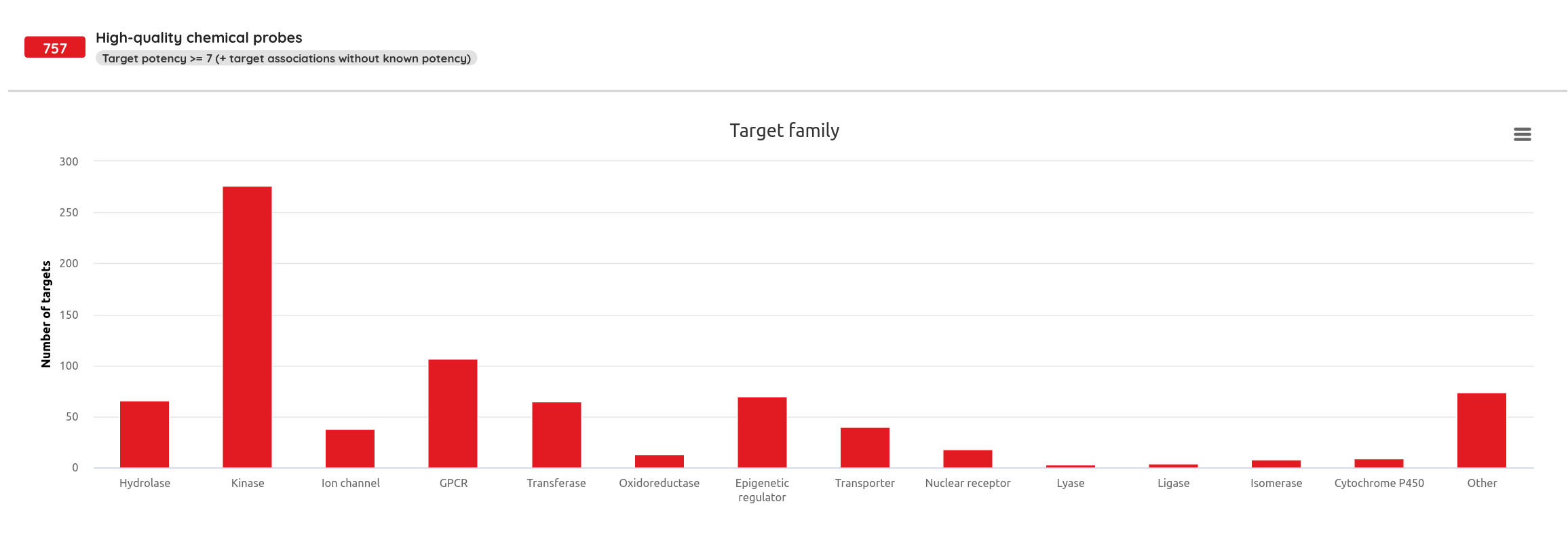
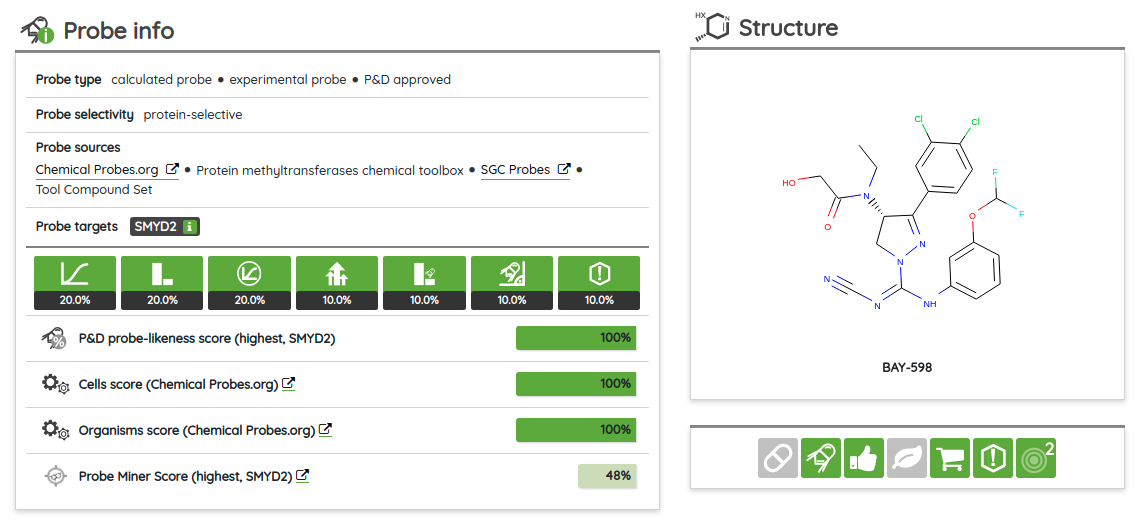
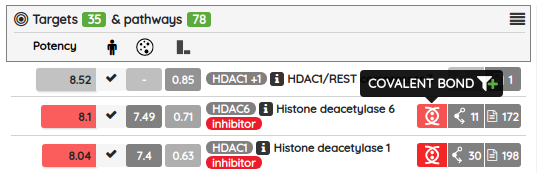
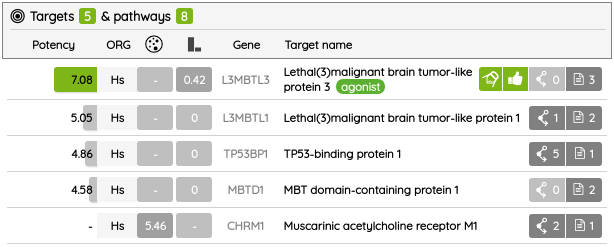
Our High-quality chemical probes set in its latest version contains 790 standardized compounds for 582 primary (directly assigned) targets.
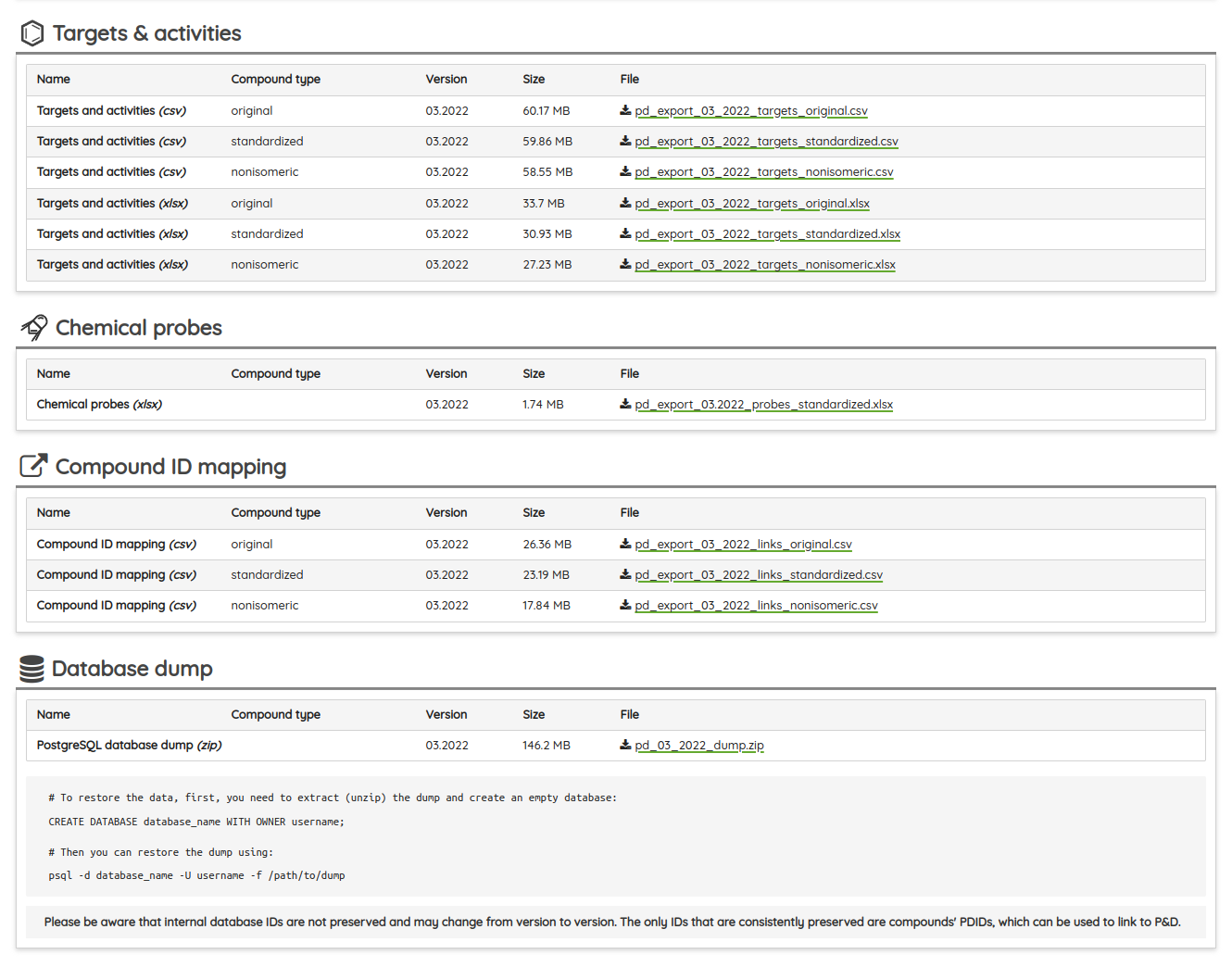
As always, you can download specialized exports as well as all data used on the portal as a PostgreSQL database dump or the SQLite database in the Download section.
We hope you'll find the new data/features useful and if you have any questions or suggestions or if you stumble upon a bug, don't hesitate to contact us through our contact form.